TÜRKIC AND EUROPEAN GENETIC DISTANCE
Reference
ROSALIND M. HARDING AND ROBERT R. SOKAL
Classification of the European language
families by genetic distance
(human variation/gene frequencies)
Proc. Nati. Acad. Sci. USA
Vol. 85, pp. 9370-9372, December 1988
Population Biology
Department of Ecology and Evolution, State
University of New York at Stony Brook, Stony Brook, NY 11794-5245
Contributed by Robert R. Sokal, August 24, 1988
INTRODUCTION
The citation below from the Proceedings of National Academy of Sciences article provides a graphic illustration of the relationship between the genetical composition of a category of population, and its linguistic classification. While they are generally presumed to mutually correlate, and that implicit or assumed correlation is laid in the foundation of some reconstructive historical hypotheses, the study explicated in the article demonstrates the effects of the linguistic conversions of entirely opposite nature.
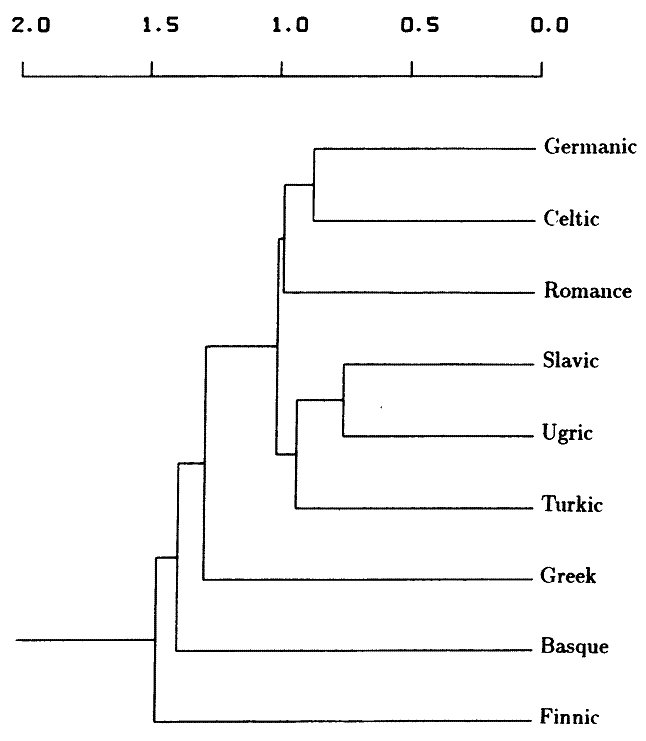
The study revealed that the peoples presently speaking Slavic, Ugric and Türkic languages constitute one genetic distinct group (Fig. 1). This is consistent with the concept that the present speakers of Slavic language genetically are predominantly Slavisized Finno-Ugrians and Turks. A further study of the Northern Slavic and Southern Slavic subdivisions in Eastern Europe would illustrate that the Northern Slavic populace is genetically connected with the Finno-Ugric group, and the Southern Slavic populace is genetically connected with the Türkic group. The carriers of the European "Indo-European" German, Celtic and Roman languages also constitute a separate distinct group (Fig. 1). Timewise, the geographical spread of the Slavic language is consistent with the advance of the Slavic colonization in the Northern and Southern parts of the Central and Eastern Europe during the last millennia. The genetic aspect of the colonization is manifested in that the "genetic distances of some interphylum language-family pairs, such as those between Slavic and Ugric speakers, or between Turkic and Ugric speakers, however, are closer than some distances within a phylum, as between Greek and Celtic speakers or between Finnic and Ugric speakers."
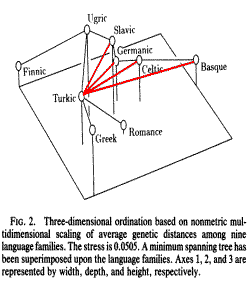
The clustering of the genetically closest Slavic-Ugric and Greek-Türkic (Fig. 2) pairs may indicate the processes of the Proto-Baltic and Proto-Greek periods, when the Baltic predecessors of the Proto-Slavs, and the later Proto-Slav splinter themselves had a close genetic connection with the Finno-Ugrian population in the Northern Europe, and Türkic population in the Southern Europe, and when the Greek predecessors and later ancient Greeks had a close genetic connection with the Türkic Scythian and later Sarmatian population in the Southern Europe.
The seemingly non-concordant genetical links should be, and for better studied cases are, detectable through the linguistic influences. Some of these cases are widely known, like the heavy layer of Türkic lexicon in the Russian language, the others have not been studied specifically or impartially, and may be helped by the indications of the genetical proximity.
Links
For a Word version click here ==>
Classification of the European language families by genetic distance
(human variation/gene frequencies)
ABSTRACT
Genetic distances among speakers of the European language families were computed by using gene frequency data for human blood group antigens, enzymes, and proteins of 26 genetic systems. Each system was represented by a different subset of 3369 localities across Europe. By subjecting the matrix of distances to numerical taxonomic procedures, we obtained a grouping of the language families of Europe by their genetic distances as contrasted with their linguistic relationships. The resulting classification largely reflects geographic propinquity rather than linguistic origins. This is evidence for the primary importance of short-range interdemic gene flow in shaping the modern gene pools of Europe. Yet, some language families-i.e., Basque, Finnic (including Lappish), and Semitic (Maltese)-have distant genetic relationships with their geographic neighbors. These results indicate that European gene pools still reflect the remote origins of some ethnic units subsumed by these major linguistic groups.
The aim of analyses of current gene frequency patterns is to infer the microevolutionary processes that have generated these patterns. Such inferences are facilitated when the investigator can employ other relevant variables in the analysis. In human populations such variables are geographic distance and language. Genetic similarity may be due to (i) geographic proximity or (ii) relationships reflected by language phylogeny. If genetic relations among languages reflect their linguistic origins, we expect strong congruence between genetic affinities and linguistic relationships. (The common origin and phylogenetic divergence of several of the language families of Europe is well established, see ref. 1.) Alternatively, if genetic affinities between language families are inversely proportional to spatial distance, they may be attributed to localized gene flow. This is Malecot's isolation by-distance model (2), which assumes stochastic divergence of populations from a common origin. The fit of these alternative models will be tested by comparing the observed genetic distances between pairs of language-family regions with (i) their spatial distances and (ii) their linguistic distances.
We prefer, with Lalouel (3), to calculate genetic distances based on a minimum of genetic assumptions. Generally, most calculations of genetic distances among human populations are highly correlated (4), giving good reason to choose the simplest computational method. However, one particular feature of the data set on which this study is based requires special attention. To examine variability on a continental scale, it was necessary to combine data from a large number of independent studies and, as a result, each genetic system is based on a different sampling scheme. Although there is some overlap in the sampling localities for different genetic systems, the final data matrix is unbalanced by the absence of observations at a given locality for various genetic systems. Thus, genetic distances had to be computed separately for each genetic system among the particular set of locality samples representing that system.
MATERIALS AND METHODS
The 12 living language families in Europe fall into five language phyla as follows (1):
Indo-European
(Albanian, Baltic, Celtic, Germanic, Greek, Romance, and
Slavic);
Finno-Ugric [Finnic and Ugric (Hungarian)];
Altaic (Turkic);
Afro-Asiatic [Semitic (Maltese)];
and
Language Isolates (Basque).
A linguistic distance matrix of language-family relationships was constructed by setting the Baltic-Slavic distance to 1 (these are the only two Indo-European families for which close genetic affinities are generally accepted, see ref. 1), all other distances between language families within a phylum to 2, and distances between language families belonging to different phyla to 4. Thus, language distances mostly contrast intraphylum and interphylum distances. A geographic distance matrix between all pairs of language families was computed from great-circle distances between subjectively chosen centers of language-family regions. Genetic distances were calculated by using frequency data for human blood antigens, enzymes, and proteins of 26 genetic systems, each for a different subset of 3369 localities across Europe. Because of the different spatial sampling for each genetic system, we computed genetic distances separately for each system. All localities were also assigned a language-family affiliation. The systems and the sources of the data are described elsewhere (5-7). Sample sizes range from 50 to many thousands of persons. Previous work (5) has shown that the simplest of these distances, that due to Prevosti et al. (8), provided essentially the same information as more elaborate formulations. It was, therefore, adopted here. To allow for possible bias due to different ranges of gene frequencies, we also standardized the distances. For each system we first calculated genetic distances for all pairs of localities and subsequently averaged over all locality pairs representing a particular pairwise combination of language families. This yielded, for a given system, an average genetic distance for each pair of language families. However, for some systems, we lacked localities to represent one or more language families and could not compute distances for certain pairwise combinations of language families. This resulted in genetic distance matrices for some systems with missing values for some pairwise comparisons. Since different genetic systems are based on different sets of localities, the particular pairwise combinations missing in the genetic distance matrices vary among systems. The final genetic distance matrix (Table 1) was obtained by averaging over all systems....
...Congruence between genetic, linguistic, and geographic distances was tested for significance as described (5). Using methods of quadratic assignment (11), we calculated pairwise Mantel matrix correlations (12, 13) and investigated three way relations between the distance matrices by computing partial correlations (14). These correlations were tested for significance by Monte Carlo permutation methods. These computations were carried out by using the R package for multivariate data analysis (15).
RESULTS
Fig. 1 shows the hierarchic clustering of genetic distances for 9 language families. A Germanic-Celtic cluster is joined later by Romance, and a Slavic-Ugric cluster is joined by Turkic. Finnic, Basque, and Greek are outliers to these clusters. Including Albanian, Baltic, and Semitic in the analysis changes the phenogram by affiliating Greek with Albanian, Baltic with Turkic, and clustering Germanic-Celtic with Slavic-Ugric before adding Romance. Semitic, Basque, and Finnic are outliers to the clusters of 12 language families.
An ordination of the genetic distance matrix (Fig. 2) depicts the relative genetic distances between the nine language families. Finnic and Basque are outliers at opposite ends of the ordinated space. The Celtic-Germanic and Slavic-Ugric language-family pairs are evident along the first axis which runs roughly East-West. The second axis approximates a North-South gradient. In the minimum spanning tree, Turkic links Ugric and Greek with Romance. The position of Romance is central on the first and second axes, but isolated by the third, explaining its variable affiliation during clustering.
Genetic distance (Gen) correlates significantly with geography (Geo) but not with language (Lan). The pairwise correlations of distance matrices based on nine language families are as follows: Gen x Geo = 0.468 (P < 0.01), Gen x Lan = 0.182 (P>0.05), and Geo x Lan = 0.177 (P>0.05). The partial correlations are (Gen x Geo)-Lan = 0.451 (P < 0.01) and (Gen x Lan)-Geo = 0.114 (P > 0.05). Geography determines 20.3% of the variance of the genetic distances, language determines only 1.0%o, and factors common to geography and language determine 1.6%.
One might have expected a high and significant Gen x Lan correlation, because speakers of a particular language (family) tend to be found settled near each other. In other words, when geographic distances between samples are small we expect linguistic distances to be small, and vice versa. If geography is likewise correlated with genetics, then genetic and linguistic distances should also be positively correlated. But the Geo x Lan correlation in this study is low because centers of language phyla are positioned in Europe both relatively close and far apart spatially causing the relationship between geography and language to break down. Therefore, the common effect of geography does not produce a high correlation between genetics and language. This finding contrasts with the significant correlation between genetic and language distances reported by Sokal (5). In that study correlations among genetics, linguistics, and geography were calculated for pairwise locality distances. Since a finer scale of distances was used, both the correlation of geography and of language and the common effect of geography were greater.
DISCUSSION
This study shows that genetic distances between the European language families do not reflect their accepted linguistic relationships. If we group the language families by their linguistic origins, there should be a cluster of the Indo- European language families, Baltic and Slavic being most closely related, a separate branch for the Finnic and Ugric speakers, and separate coordinate branches for the Turkic, Semitic, and Basque language families. The genetic distances of some interphylum language-family pairs, such as those between Slavic and Ugric speakers, or between Turkic and Ugric speakers, however, are closer than some distances within a phylum, as between Greek and Celtic speakers or between Finnic and Ugric speakers. The low matrix correlation between genetics and language confirms the lack of agreement between presumed language phylogeny and the observed genetic distances.
If genetic distances reflect geographic proximity, we should be able to predict genetic affinity from a clustering of the great-circle distances between language families. This yields Germanic-Celtic and Romance-Basque as mutually closest pairs, with the two pairs together forming a major cluster. Likewise, Slavic-Ugric and Greek-Turkic both cluster as pairs. Finally, Finnic is an outlier. Clustering all 12 language families by great-circle distance enlarges the Greek- Turkic cluster into one that also includes Albanian and Semitic and places Baltic-Finnic as an outlying pair. Considerable concordance between geographic proximity and observed genetic relations is evident and confirmed by the significant correlation between geographic and genetic distances. The role of geography can also be seen in the ordinations. Disregarding the outliers, Fig. 2 demonstrates an East-West separation of geographically adjacent language family pairs, Germanic-Celtic from Turkic-Greek with Slavic- Ugric intermediate. The small Greek-Albanian genetic distance is also consistent with geographic proximity, although we attach less confidence to this value. The observations made here are supported by a largely geographic clustering of European map quadrats characterized by gene frequencies (16).
A geographic gene flow model does not, however, explain why the Basque and Finnic language families are outliers both in the ordinations and phenograms, nor why Semitic is an outlier in the extended dataset. These results reflect the distant origins of speakers of these language groups. The Finnic language family is given its unique genetic profile by inclusion of the Lapps. These populations, ethnically different from other Finnic speakers, apparently migrated to northern Scandinavia from northern Eurasia (17). The Basques have long been an isolated enclave, presumably descended from the pre-Indo-European inhabitants of Europe (18, 19). The Semitic speakers have North African origins. These results suggest that some modification of the strictly geographic gene flow model by language origin may provide greater concordance with the genetic relationships between language families.
We conclude that affinities between modern European gene pools have been formed primarily by relatively shortrange gene flow between geographically adjacent populations. Yet, between the speakers of some language families and their geographic neighbors, there are genetic differences that apparently reflect their remote historical and linguistic origins.
Table 1. Average genetic distance matrix among language families
|
Average genetic distance |
||||||||||||
|
|
Germanic |
Romance |
Slavic |
Finnic |
Ugric |
Greek |
Celtic |
Basque |
Turkic |
Baltic |
Albanian |
Semitic |
| Germanic | 0.789 | |||||||||||
| Romance | 0.984 | 0.895 | ||||||||||
Slavic |
0.873 | 1.006 | 0.759 |
|
|
|
||||||
Finnic |
1.353 | 1.392 | 1.368 | 1.139 |
|
|
|
|||||
Ugric |
0.912 | 1.059 | 0.746 | 1.319 | 0.679 |
|
|
|||||
|
Greek |
1.374 | 1.259 | 1.274 | 1.621 | 1.276 | 1.130 |
|
|||||
Celtic |
0.869 | 0.979 | 0.906 | 1.452 | 1.052 | 1.456 |
0.874 |
|
||||
|
Basque |
1.192 | 1.274 | 1.260 | 1.846 | 1.495 | 1.717 |
1.147 | 0.740 |
|
|||
|
Turkic |
1.081 | 0.949 | 0.963 | 1.343 | 0.887 |
1.028 |
1.193 | 1.574 | 0.852 |
|
||
Baltic |
1.025 | 1.107 | 0.842 | 1.178 | 0.741 | 0.811 |
2.049 | 1.449 | 0.719 | 0.864 |
||
Albanian |
1.223 | 1.046 | 1.035 | 1.451 | 1.216 | 0.551 |
1.670 | 2.692 | 0.643 | 0.636 |
0.383 | |
|
Semitic |
1.324 | 1.219 | 1.380 | 1.838 | 1.241 | 1.267 |
1.201 | 1.427 | 1.077 | - |
- | - |
|
Entries in the table are standardized Prevosti distances, averaged particular pairwise combination of language families are unavailable. |
||||||||||||
|
Average genetic distance from
Turkic group |
|
| Linguistic Group | Linguistic Group |
| Turkic | 0.0 |
| Celtic | 0.017 |
| Slavic | 0.021 |
| Ugric | 0.060 |
| Germanic | 0.063 |
| Romance | 0.132 |
| Basque | 0.340 |
| Baltic | 0.371 |
| Albanian | 0.371 |
| Semitic | 0.472 |
| Finnic | 0.501 |
| Greek | 0.522 |
| Averaged particular pairwise combination of language families are unavailable. | |
FIG. 1. Phenogram based on average unweighted pair-group clustering of average genetic distances among nine language families. Abscissa is average genetic distance. The cophenetic correlation coefficient is 0.842.
| Original |
Cophenetically equivalent simplified
graph (cophenetic establishes the relative point of divergence, allowing alternate tree geometries) |
 |
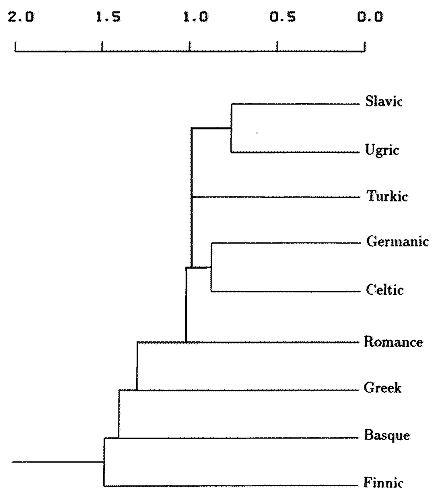 |
FIG. 2. Three-dimensional ordination based on non-metric multidimensional scaling
of average genetic distances among nine language families. The stress is 0.0505.
A minimum spanning tree has been superimposed upon the language families. Axes
1, 2, and 3 are represented by width, depth, and height, respectively.
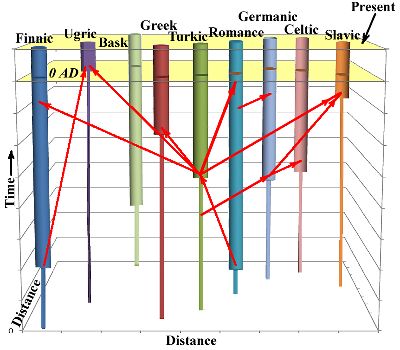
(Graph on the right illustrates relative temporal relations, thin line =
proto-language time, same scale)
1. Ruhlen, M. (1987) A Guide to the World's Languages (Stanford Univ. Press, Stanford, CA), Vol. 1.
2. Maldcot, G. (1948) Les Mathematiques de l'Hewredite6 (Masson et Cie, Paris) [Maldcot, G., trans. (1969)
The Mathematics of Heredity (Freeman, San Francisco)].
3. Lalouel, J.-M. (1980) in Current Developments in Anthropological Genetics, eds. Mielke, J. H. & Crawford, M.
H. (Plenum, New York), Vol. 1, pp. 209-250.
4. Jorde, L. B. (1985) Annu. Rev. Anthropol. 14, 343-373.
5. Sokal, R. R. (1988) Proc. NatI. Acad. Sci. USA 85, 1722-1726.
6. Sokal, R. R., Oden, N. L. & Thomson, B. A. (1988) Am. J. Phys. Anthropol. 76, 337-361.
7. Sokal, R. R., Oden, N. L., Legendre, P., Fortin, M. J., Kim, J. & Vaudor, A. (1988) Am. J. Phys. Anthropol., in press.
8. Prevosti, A., Ocana, J. & Alonso, G. (1975) Theor. Appl. Genet. 45, 231-241.
9. Sneath, P. H. A. & Sokal, R. R. (1973) Numerical Taxonomy (Freeman, San Francisco).
10. Rohlf, F. J. (1985) Numerical Taxonomy System of Multivariate Statistical Programs, Technical Report (State
Univ. of New York, Stony Brook).
11. Hubert, L. (1987) Assignment Methods in Combinatorial Data Analysis (Dekker, New York).
12. Mantel, N. (1967) Cancer Res. 27, 209-220.
13. Sokal, R. R. (1979) Syst. Zool. 28, 227-231.
14. Smouse, P. E., Long, J. C. & Sokal, R. R. (1986) Syst. Zool. 35, 627-632.
15. Legendre, P. (1985) The R Package for Multivariate Data Analysis, Technical Report (Universitd de Montreal, Montreal).
16. Derish, P. A. & Sokal, R. R. (1988) Hum. Biol., 60, 801-824.
17. Bunak, V. V. (1976) in Rassengeschichte der Menschheit, ed. Schwidetzky, I. (Oldenbourg, Munich), Vol. 4, pp. 7-101.
18. Allieres, J. (1986) Les Basques (Univ. of France Press, Paris).
19. Cavalli-Sforza, L. L. (1987) The Basque Population and Ancient Migrations in Europe, Second World Basque Congress, in press.